Add pipelines
The Launchpad lists the preconfigured Nextflow pipelines that can be executed on the compute environments in your workspace.
Platform offers two methods to import pipelines to your workspace Launchpad — directly from Seqera Pipelines, or manually via Add pipeline in Platform. This page illustrates each method, using nf-core/rnaseq as an example.
Import nf-core/rnaseq from Seqera Pipelines
Seqera Pipelines is an open-source resource that provides a curated collection of high-quality, open-source pipelines. Each pipeline includes a curated test dataset to use in a test run in just a few steps.
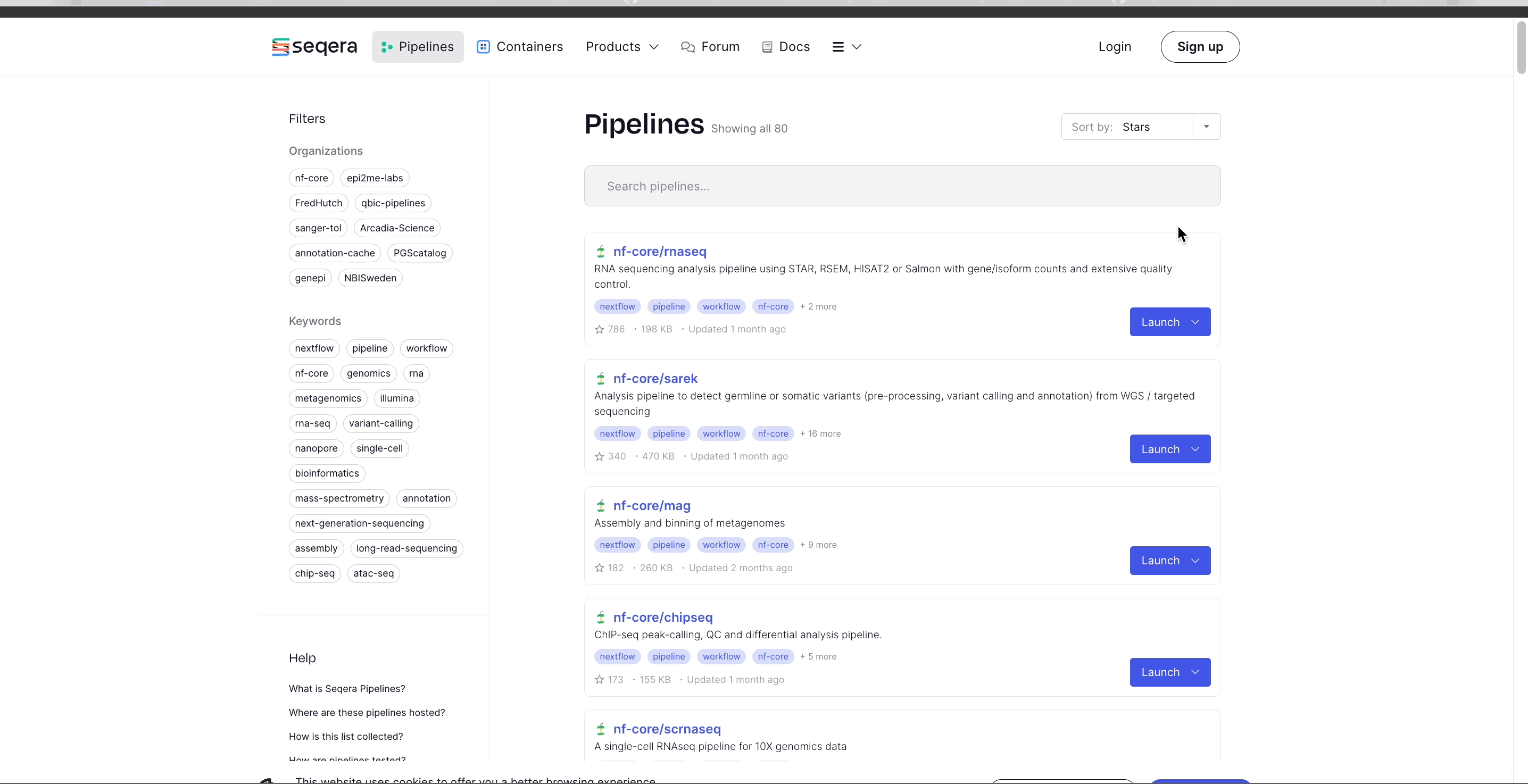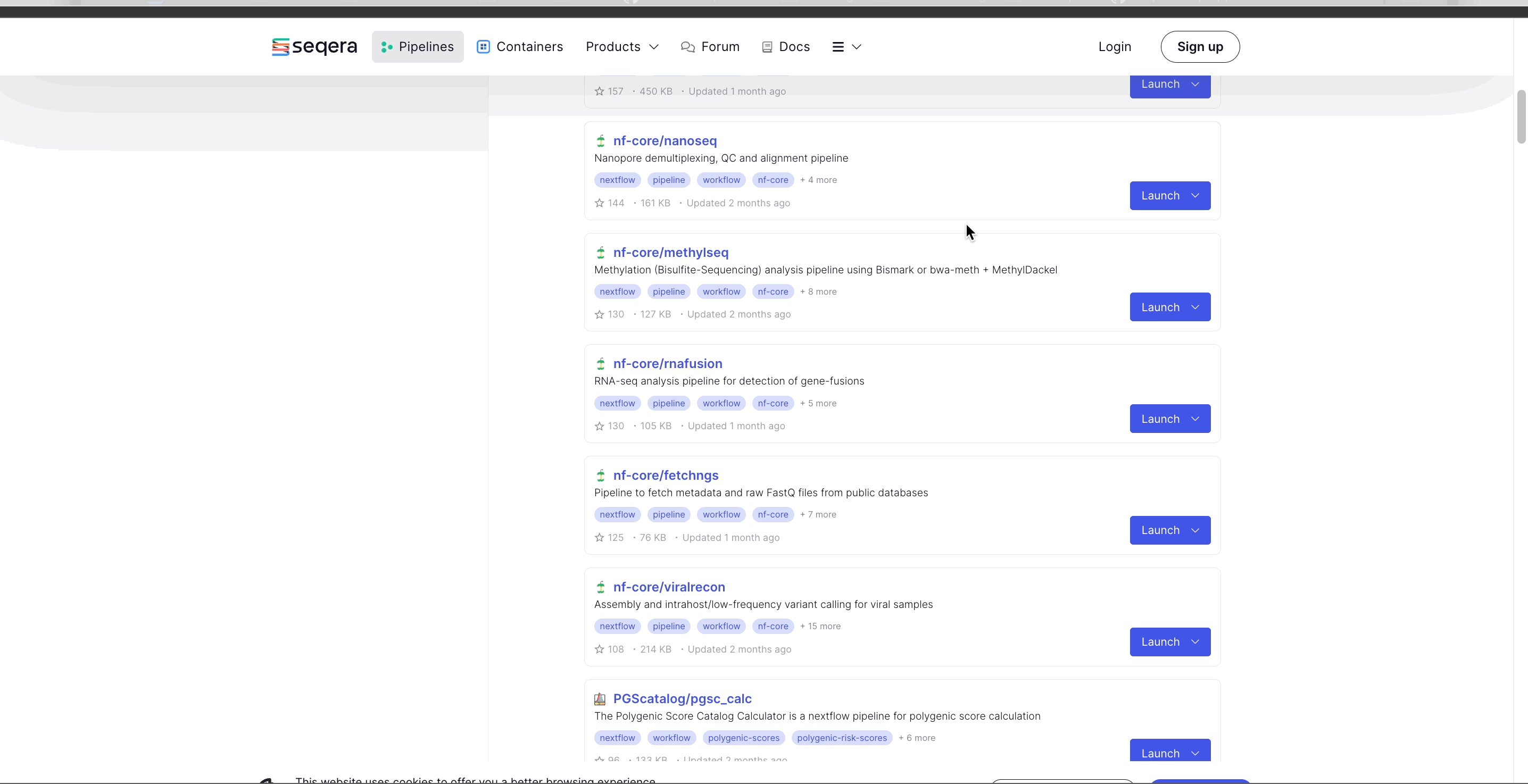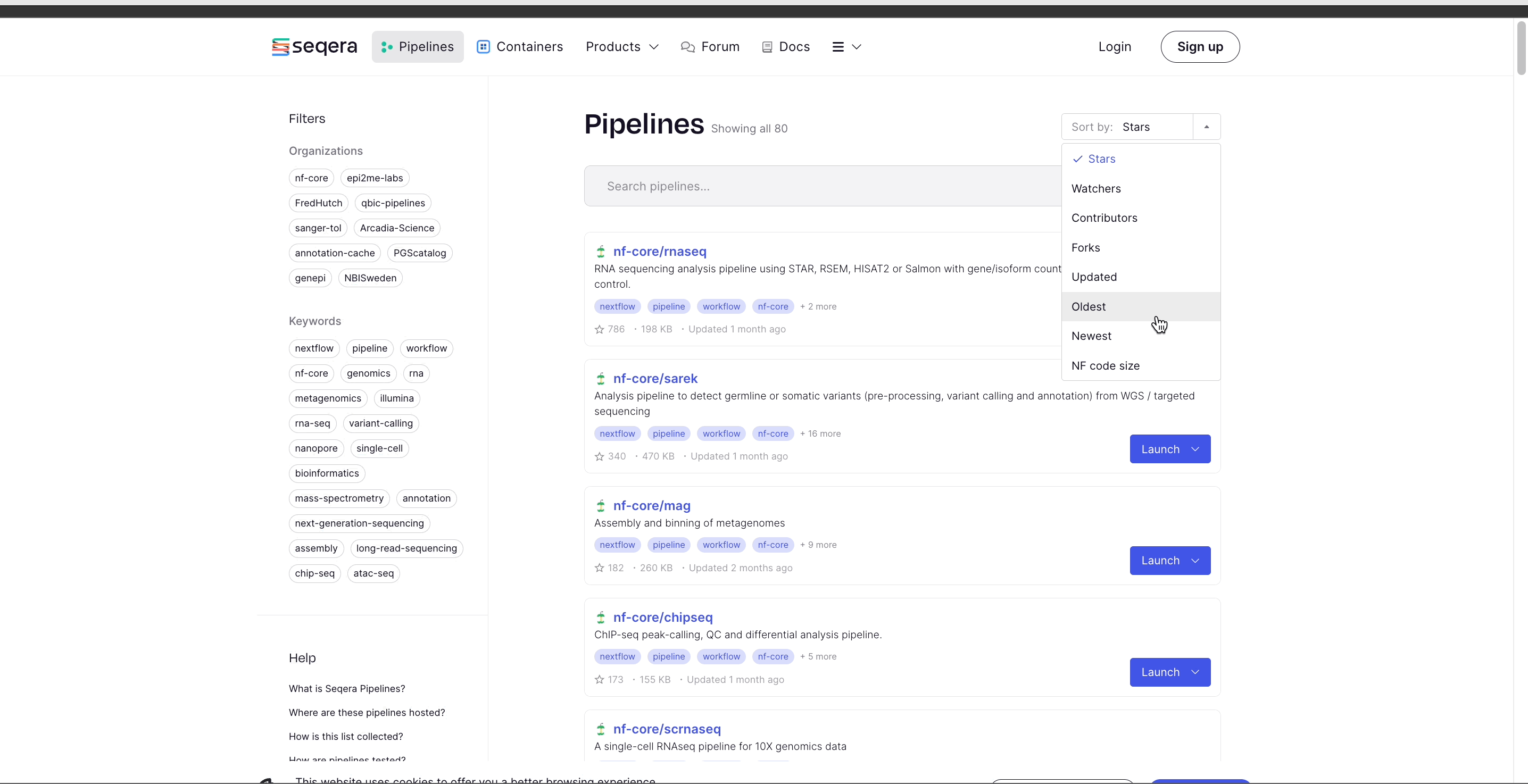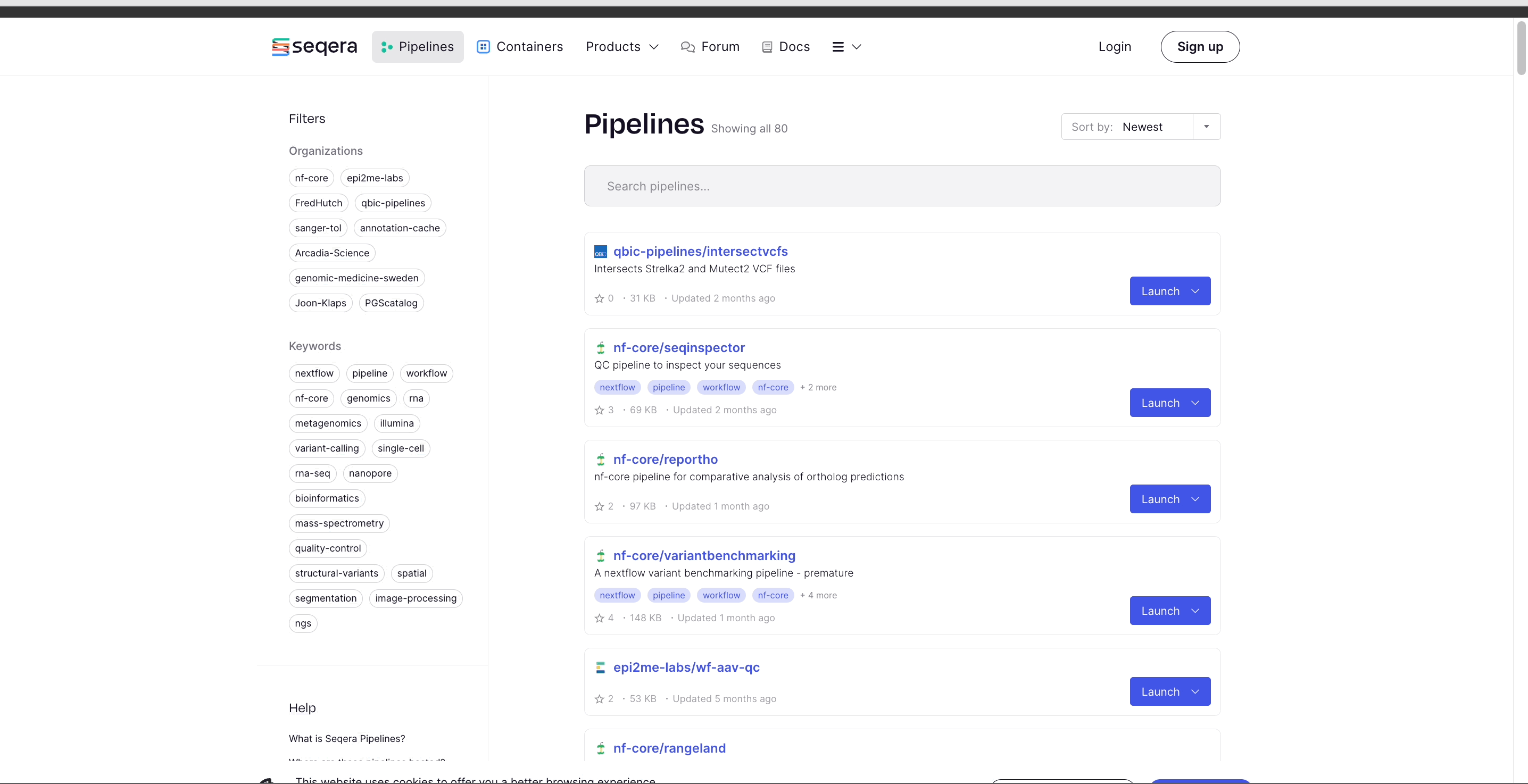
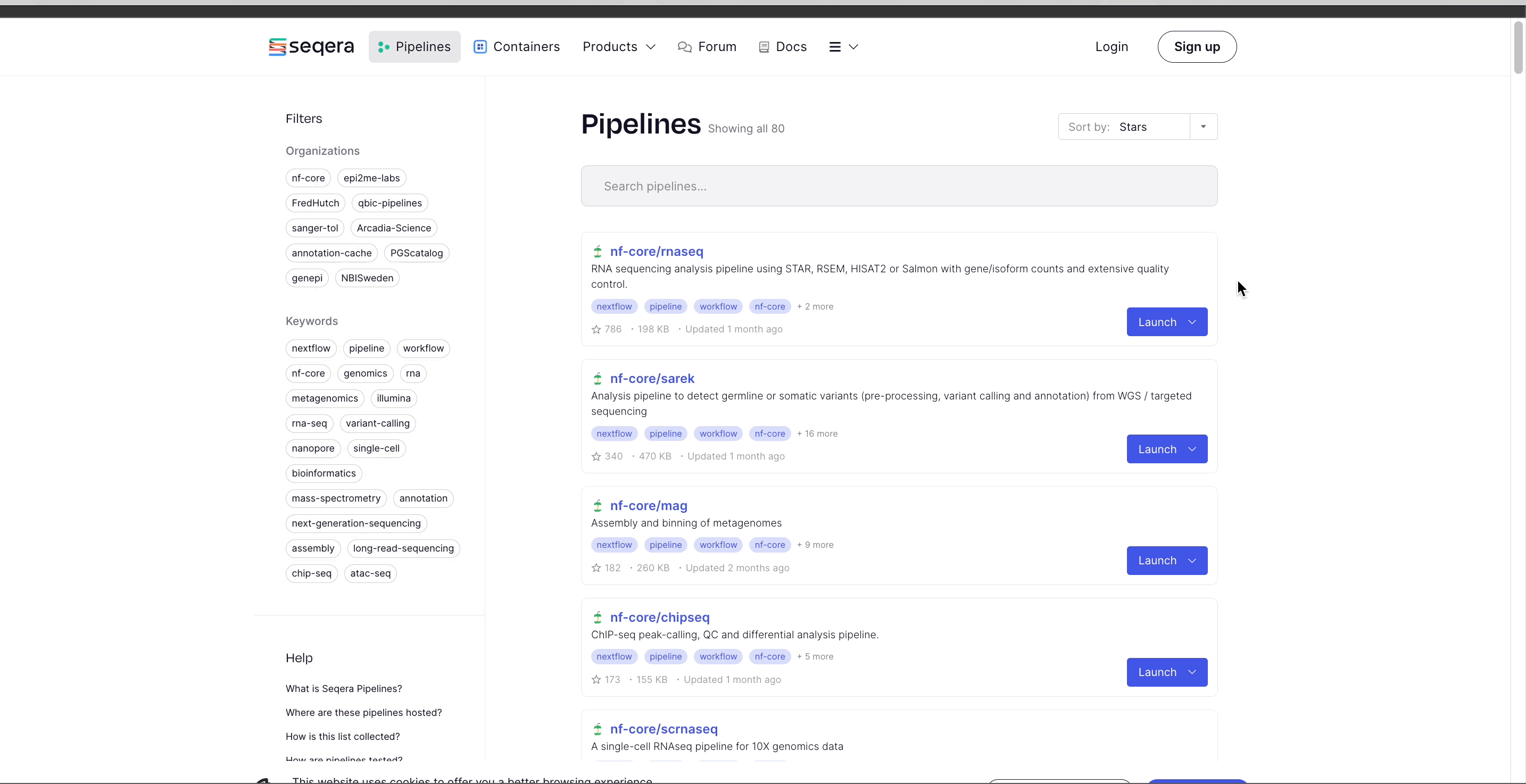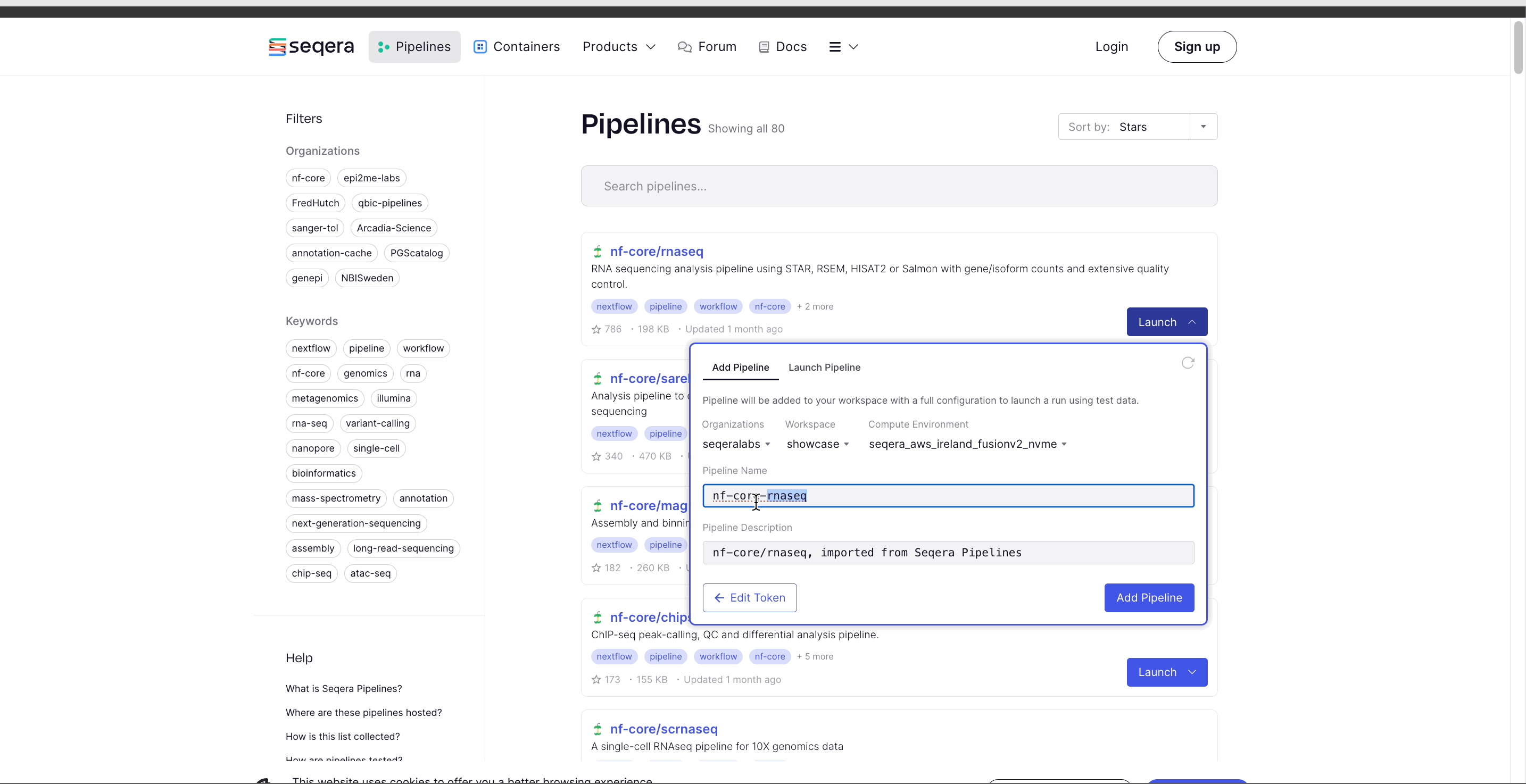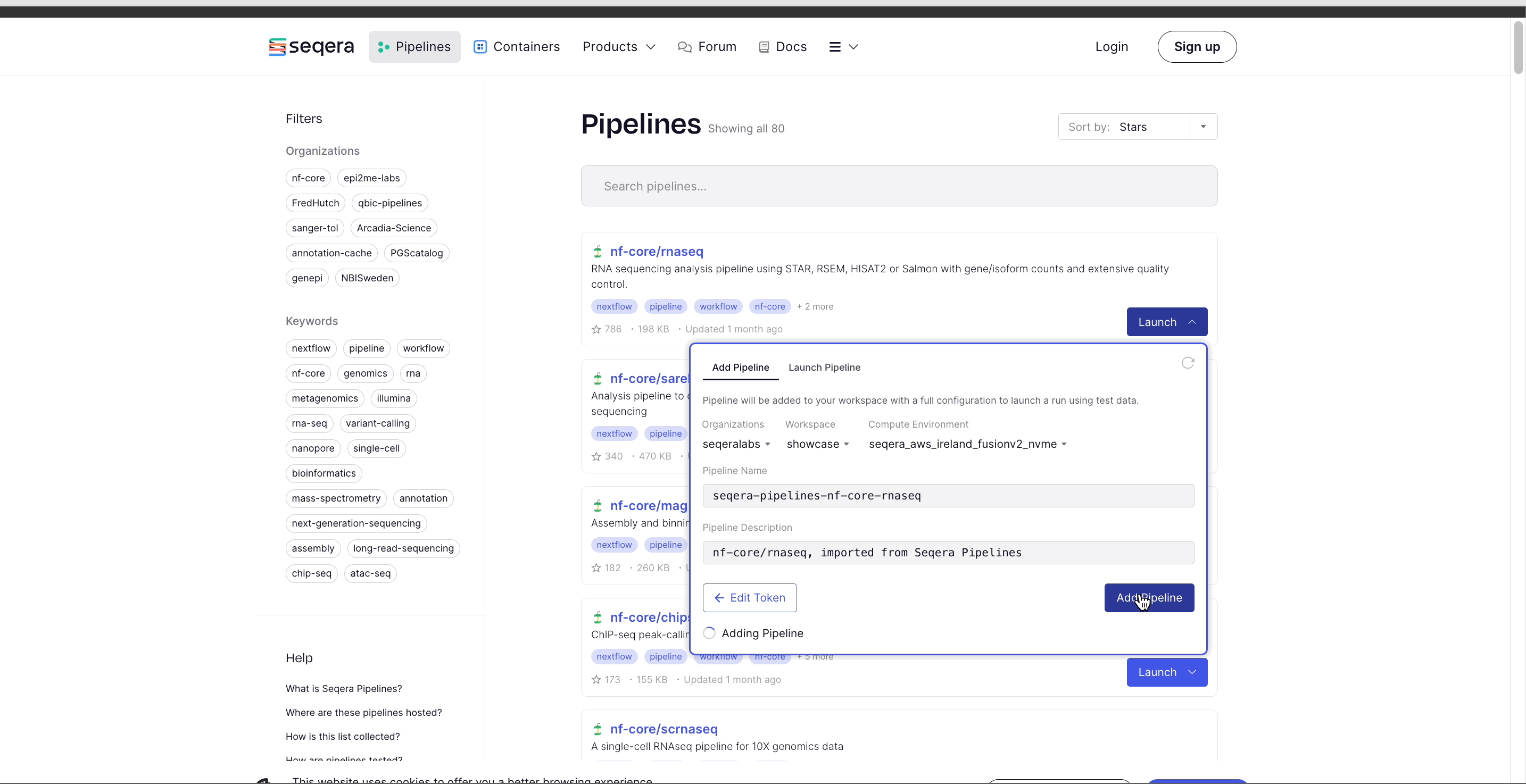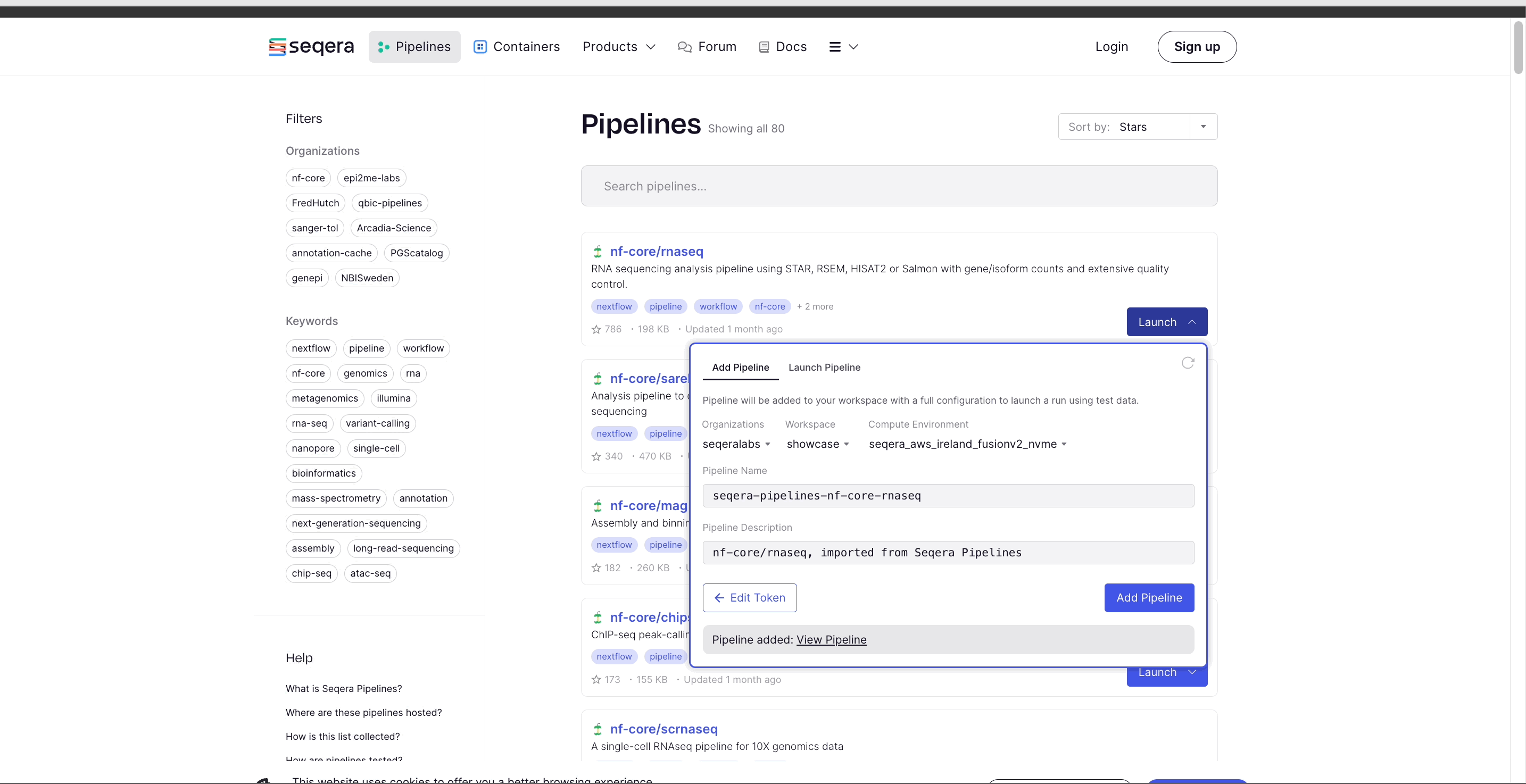
To import the nf-core/rnaseq pipeline:
- Select Launch next to the pipeline name in the list. In the Add pipeline tab, select Cloud or Enterprise depending on your Platform account type, then provide the information needed for Seqera Pipelines to access your Platform instance:
- Seqera Cloud: Paste your Platform Access token and select Next.
- Seqera Enterprise: Specify the Seqera Platform URL (hostname) and Base API URL for your Enterprise instance, then paste your Platform Access token and select Next.
If you do not have a Platform access token, select Get your access token from Seqera Platform to open the Access tokens page in a new browser window.
- Select the Platform Organization, Workspace, and Compute environment for the imported pipeline.
- (Optional) Customize the Pipeline Name and Pipeline Description.
- Select Add Pipeline.
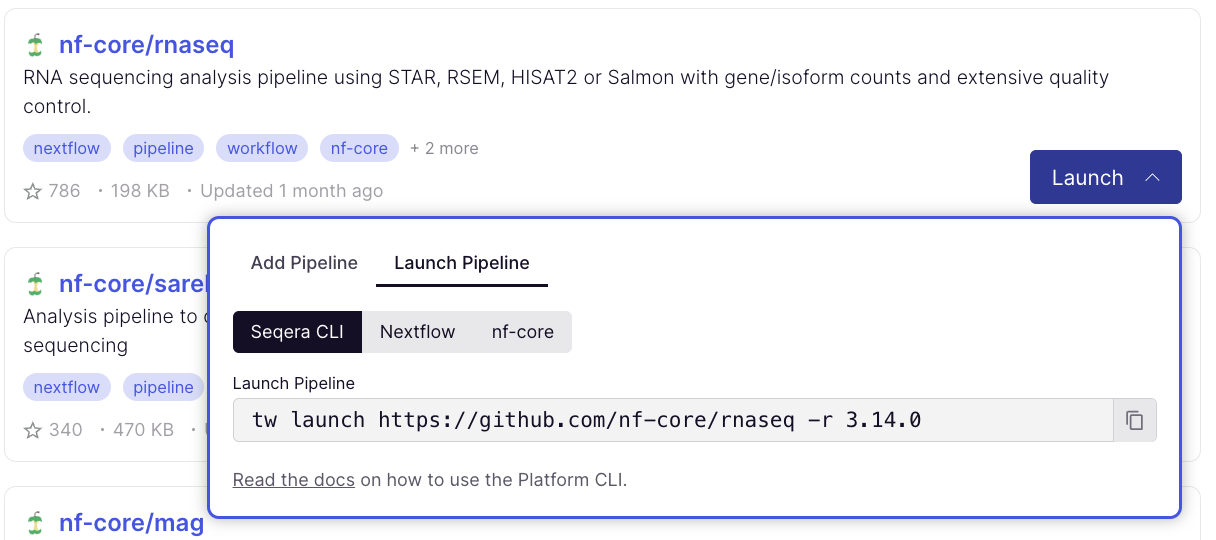
To launch pipelines directly with CLI tools, select the Launch Pipeline tab to grab commands for Nextflow, Seqera Platform CLI, and nf-core/tools:
Add nf-core/rnaseq manually
From your workspace Launchpad, select Add Pipeline and specify the following pipeline details:
- Name:
nf-core/rnaseq, or a custom name of your choice. - (Optional) Description: A summary of the pipeline or any information that may be useful to workspace participants when selecting a pipeline to launch.
- (Optional) Labels: Categorize the pipeline according to arbitrary criteria (such as reference genome version) that may help workspace participants to select the appropriate pipeline for their analysis.
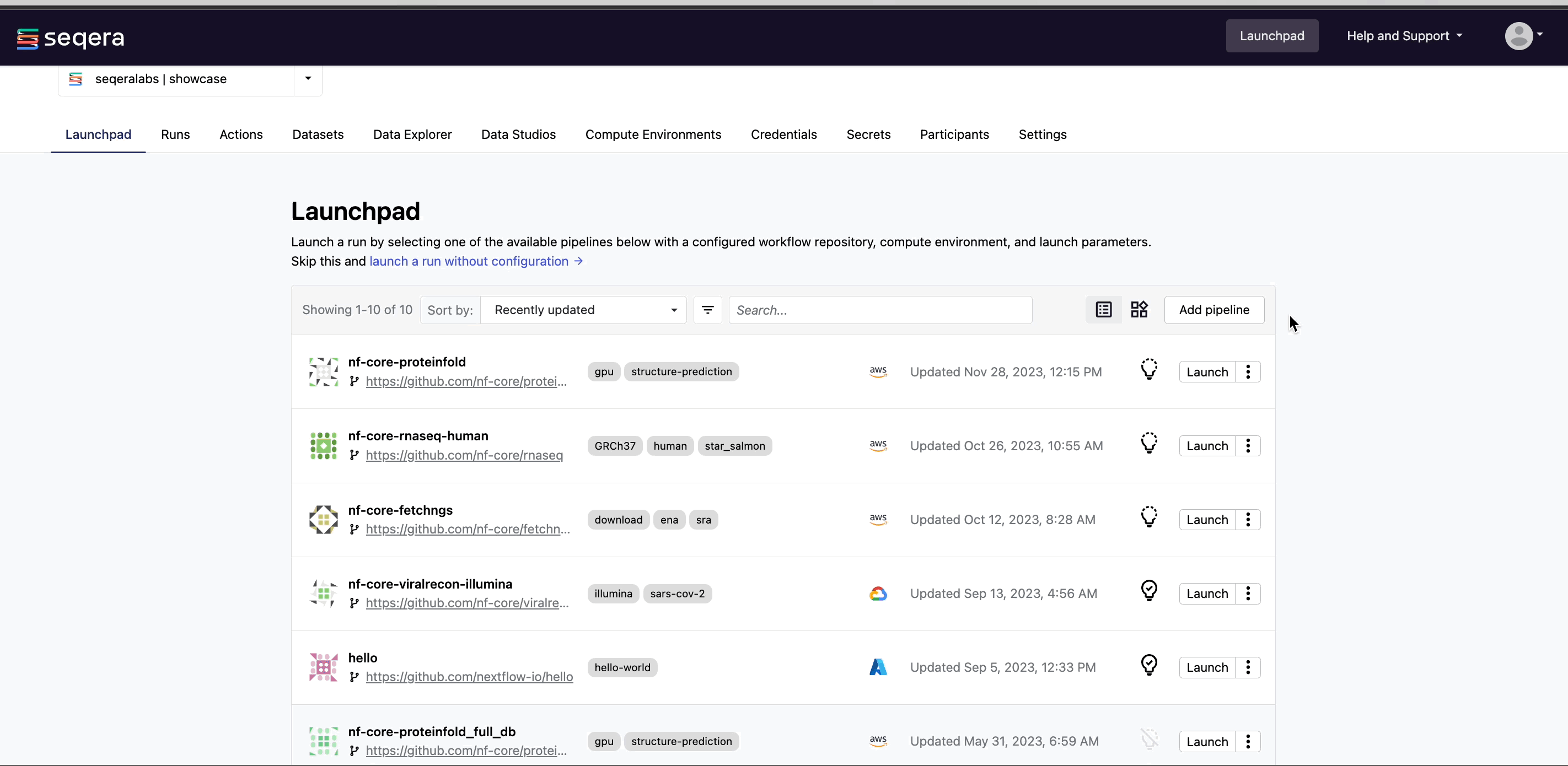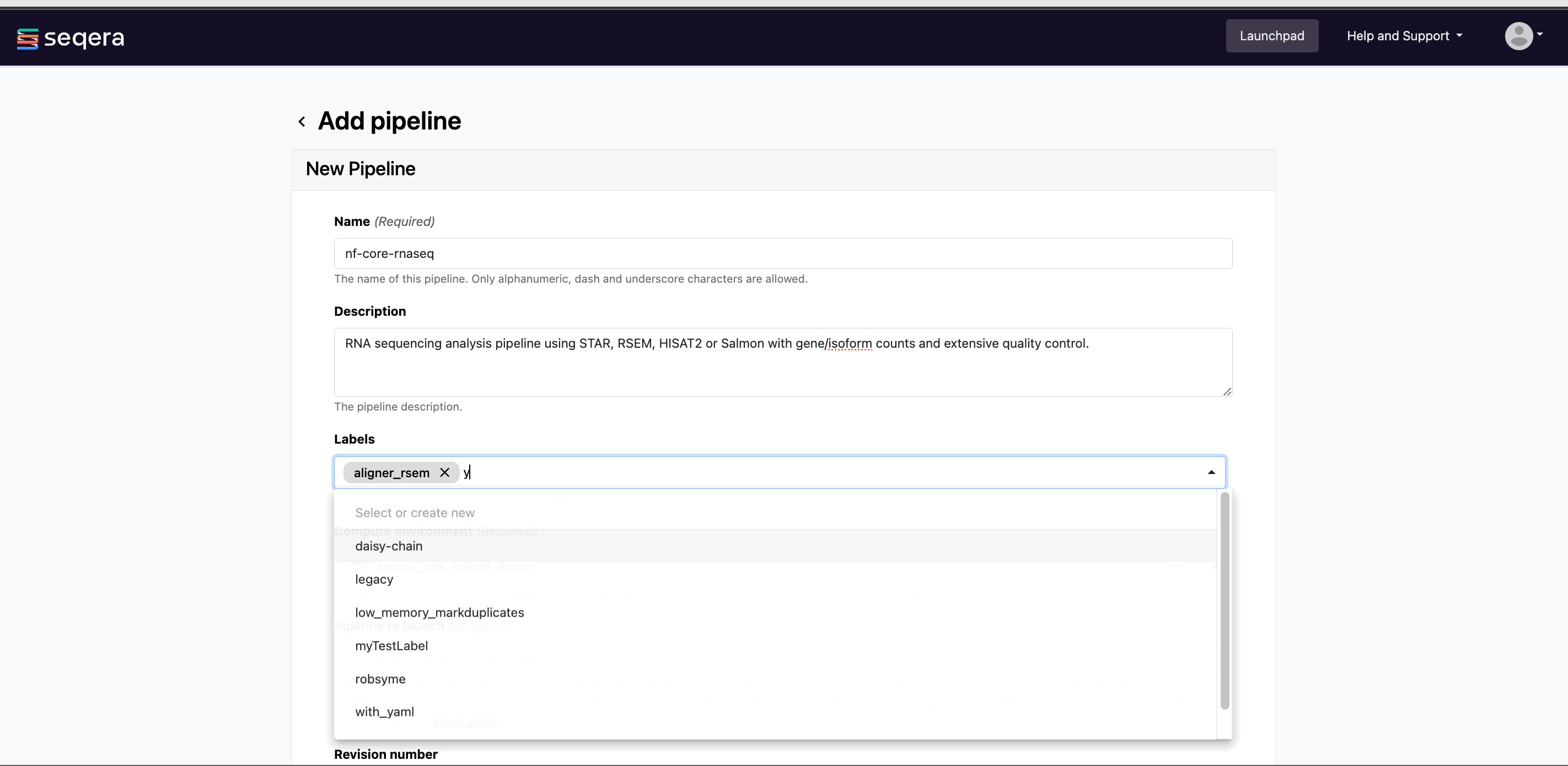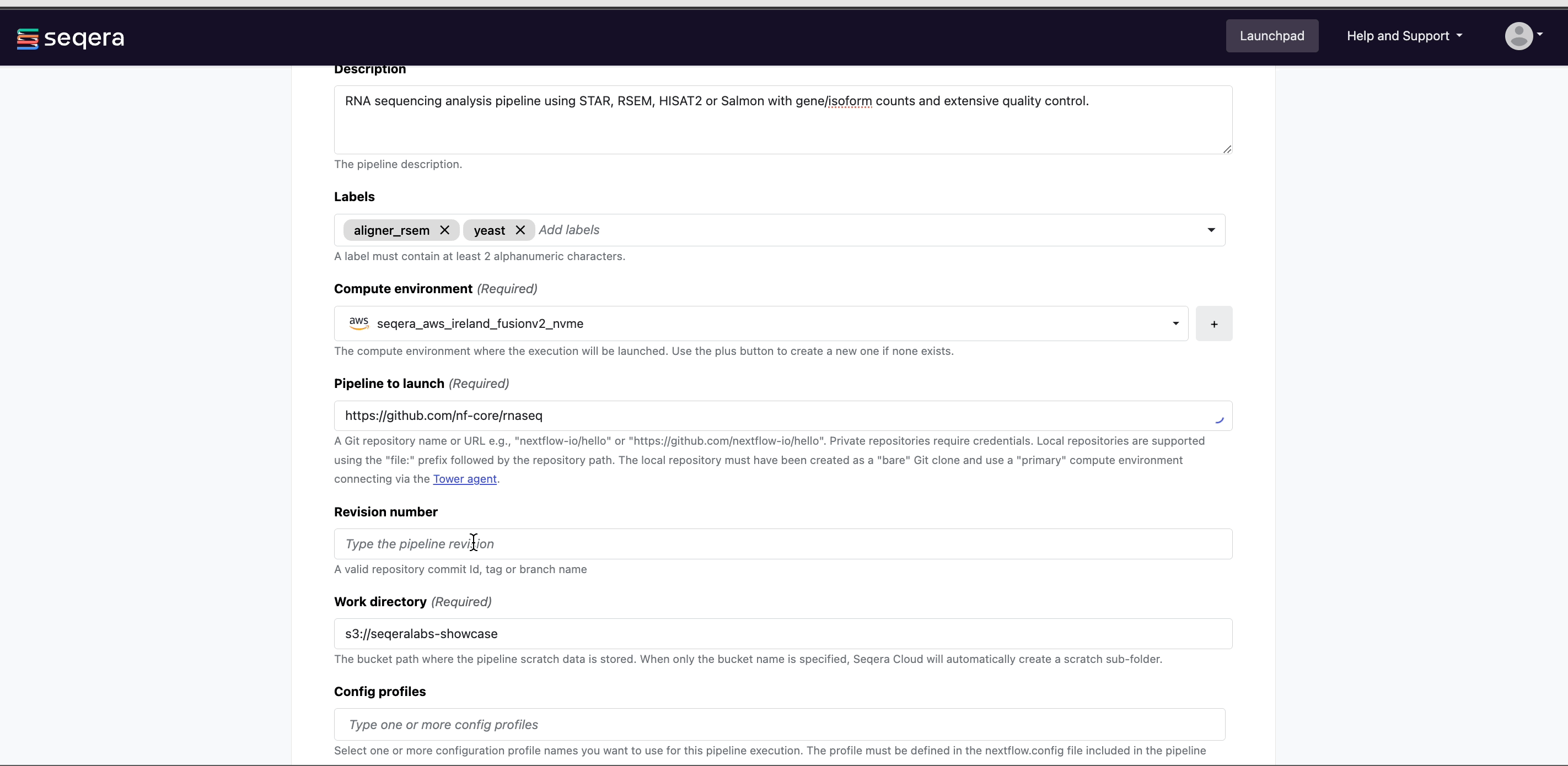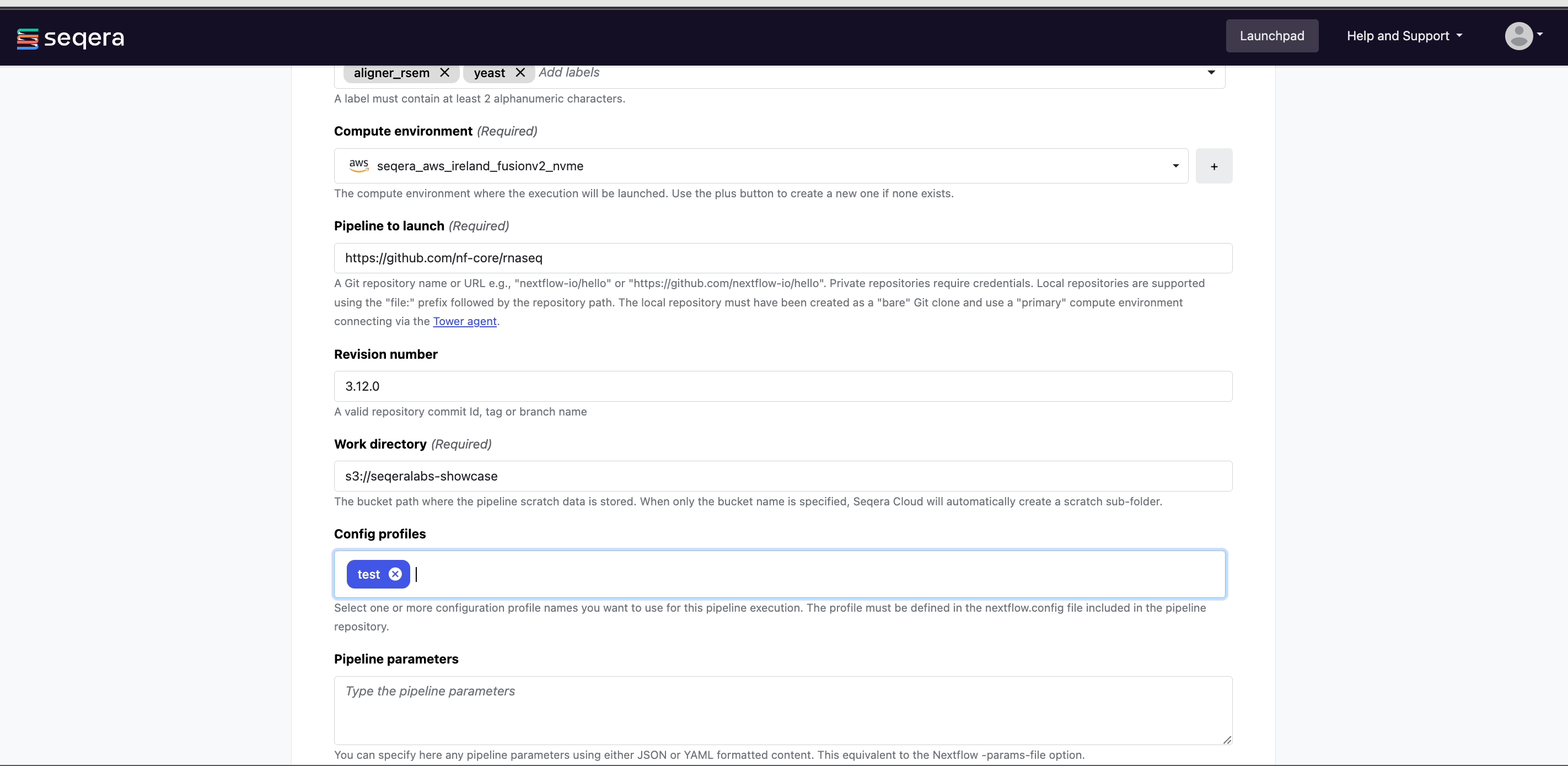
- Compute environment: Select an existing workspace compute environment.
- Pipeline to launch:
https://github.com/nf-core/rnaseq- Platform allows you to select any public or private Git repository that contains Nextflow source code.
- Revision number: Platform will search all of the available tags and branches in the provided pipeline repository and render a dropdown to select the appropriate version.
Selecting a specific pipeline version is important for reproducibility as this ensures that each run with the same input data will generate the same results.
- (Optional) Config profiles:
test- Select a predefined profile for the Nextflow pipeline.
All nf-core pipelines include a
testprofile that is associated with a minimal test dataset. This profile runs the pipeline with heavily sub-sampled input data for the purposes of CI/CD and to quickly confirm that the pipeline runs on your infrastructure. - (Optional) Pipeline parameters:
- Set any custom pipeline parameters that will be prepopulated when users launch the pipeline from the Launchpad. For example, set the path to local reference genomes so users don't have to worry about locating these files when launching the pipeline.
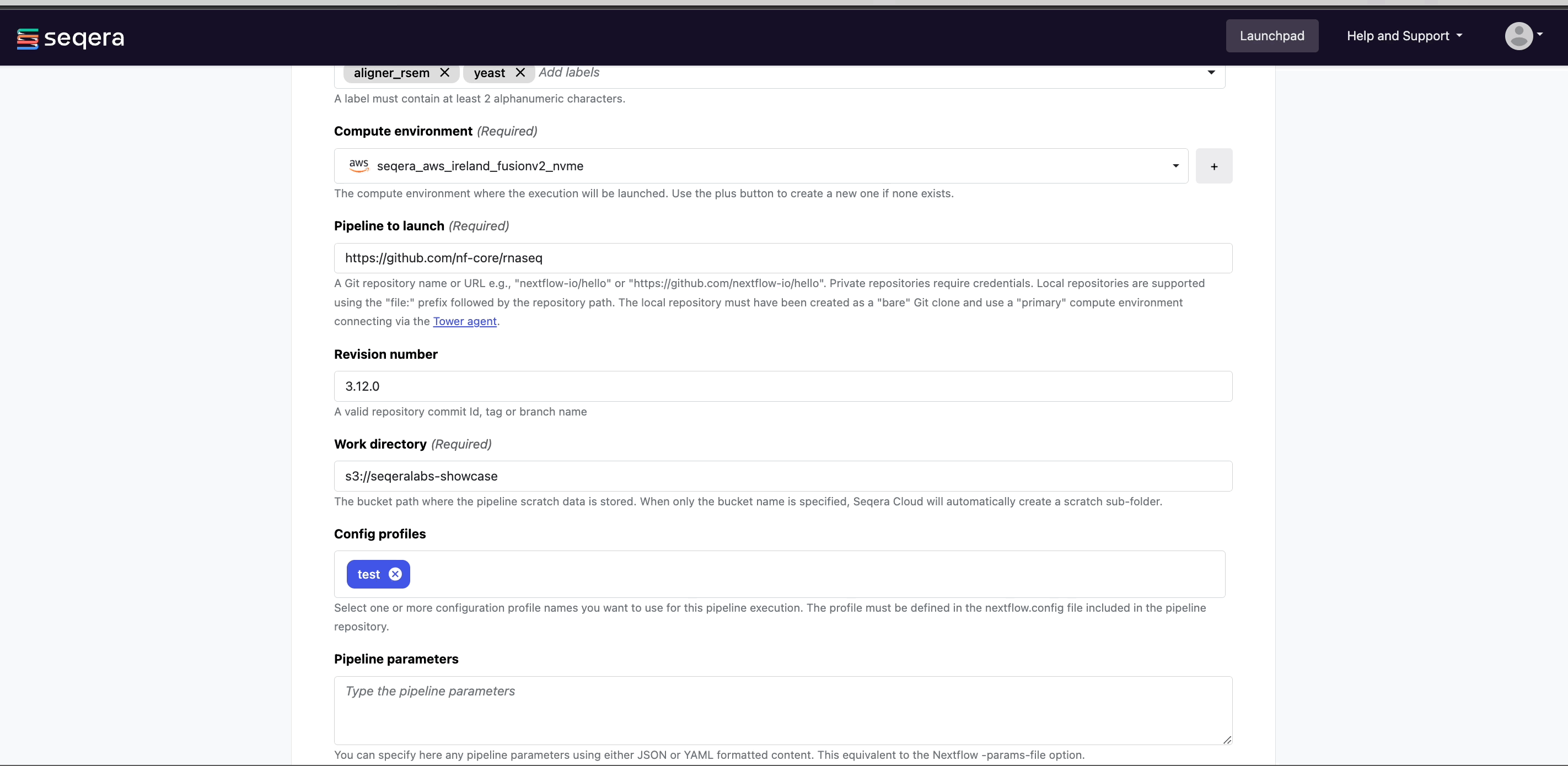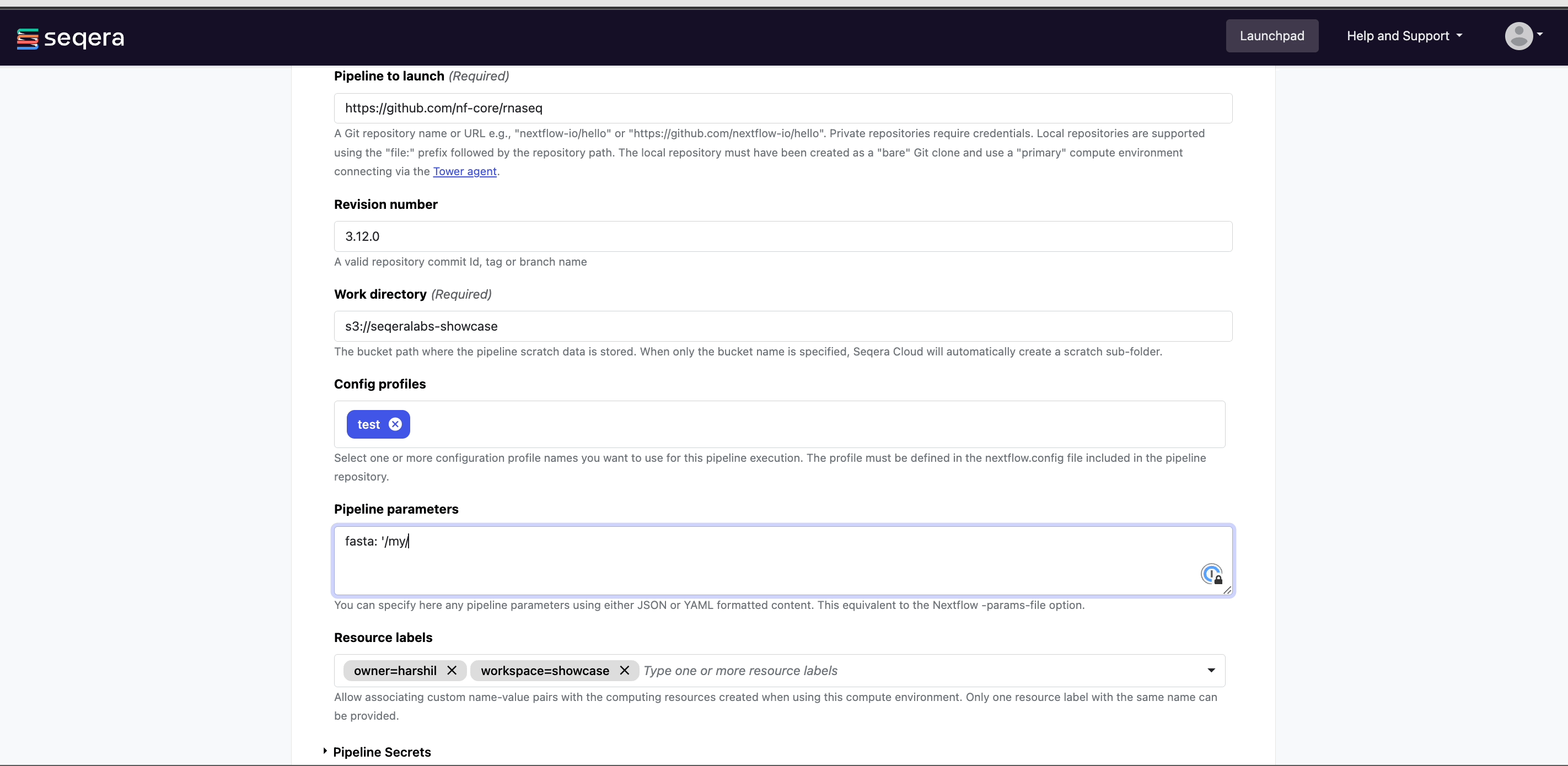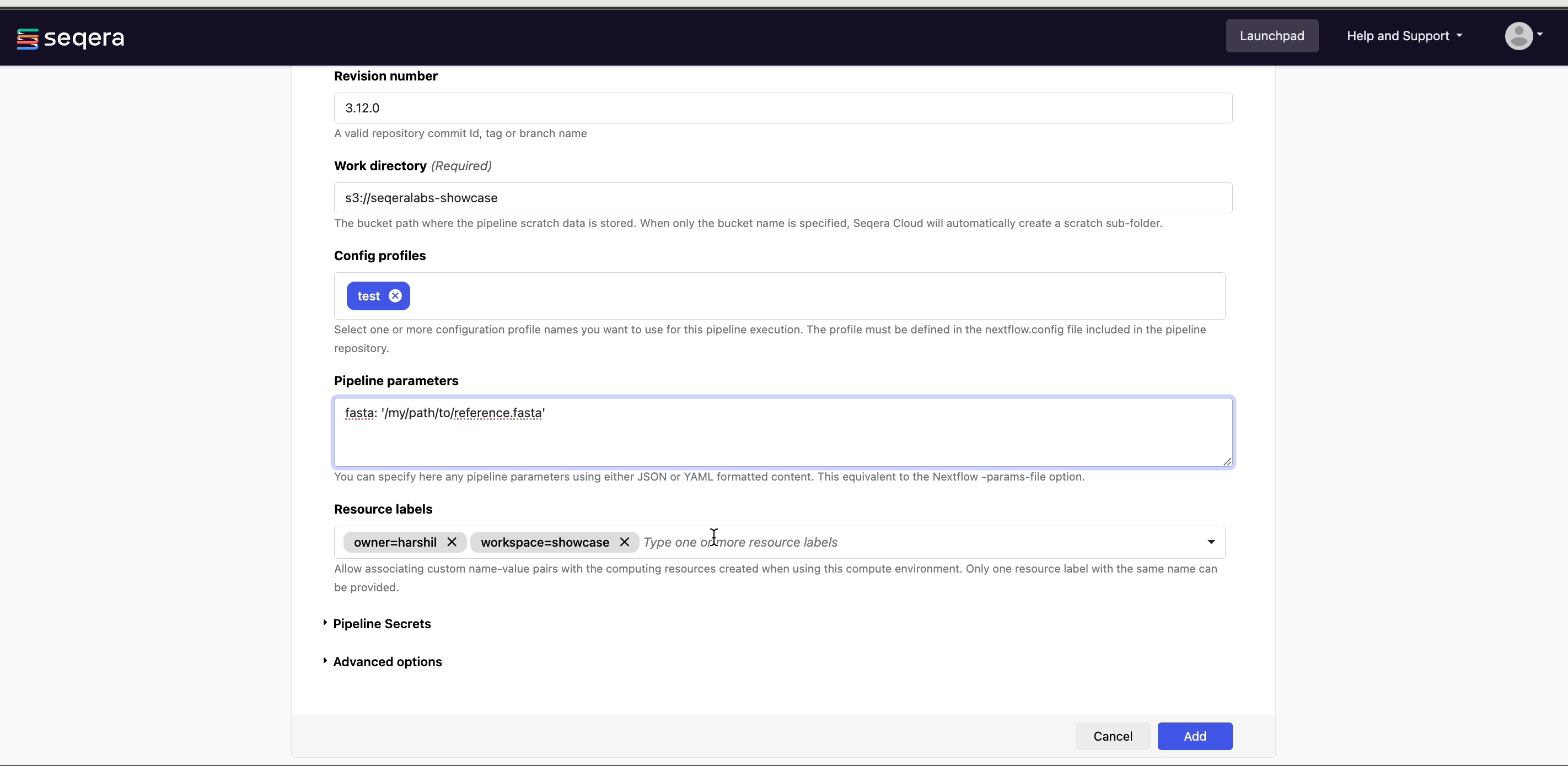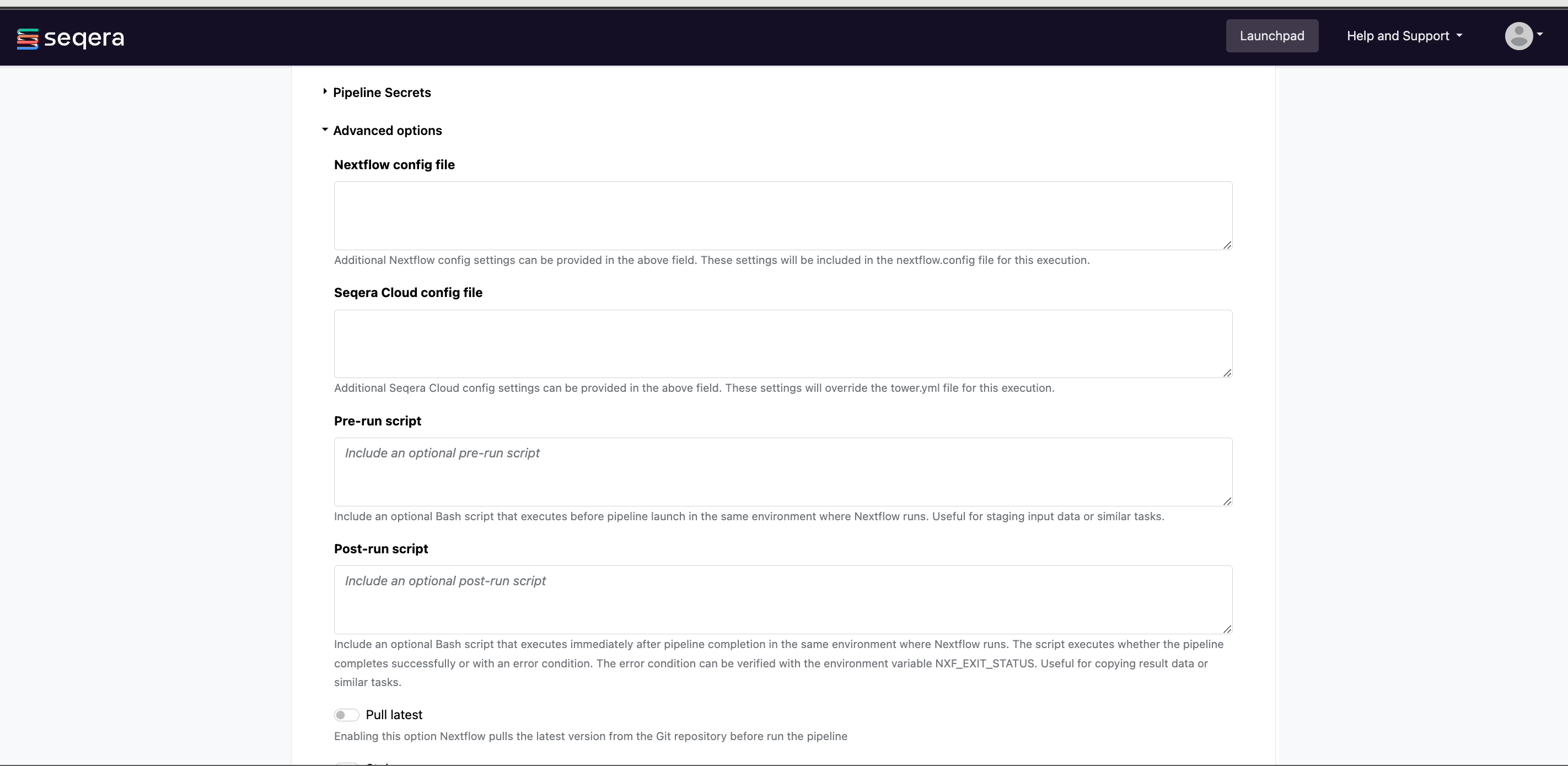
- Set any custom pipeline parameters that will be prepopulated when users launch the pipeline from the Launchpad. For example, set the path to local reference genomes so users don't have to worry about locating these files when launching the pipeline.
- (Optional) Pre-run script:
- Define Bash code that executes before the pipeline launches in the same environment where Nextflow runs.
Pre-run scripts are useful for defining executor settings, troubleshooting, and defining a specific version of Nextflow with the
NXF_VERenvironment variable.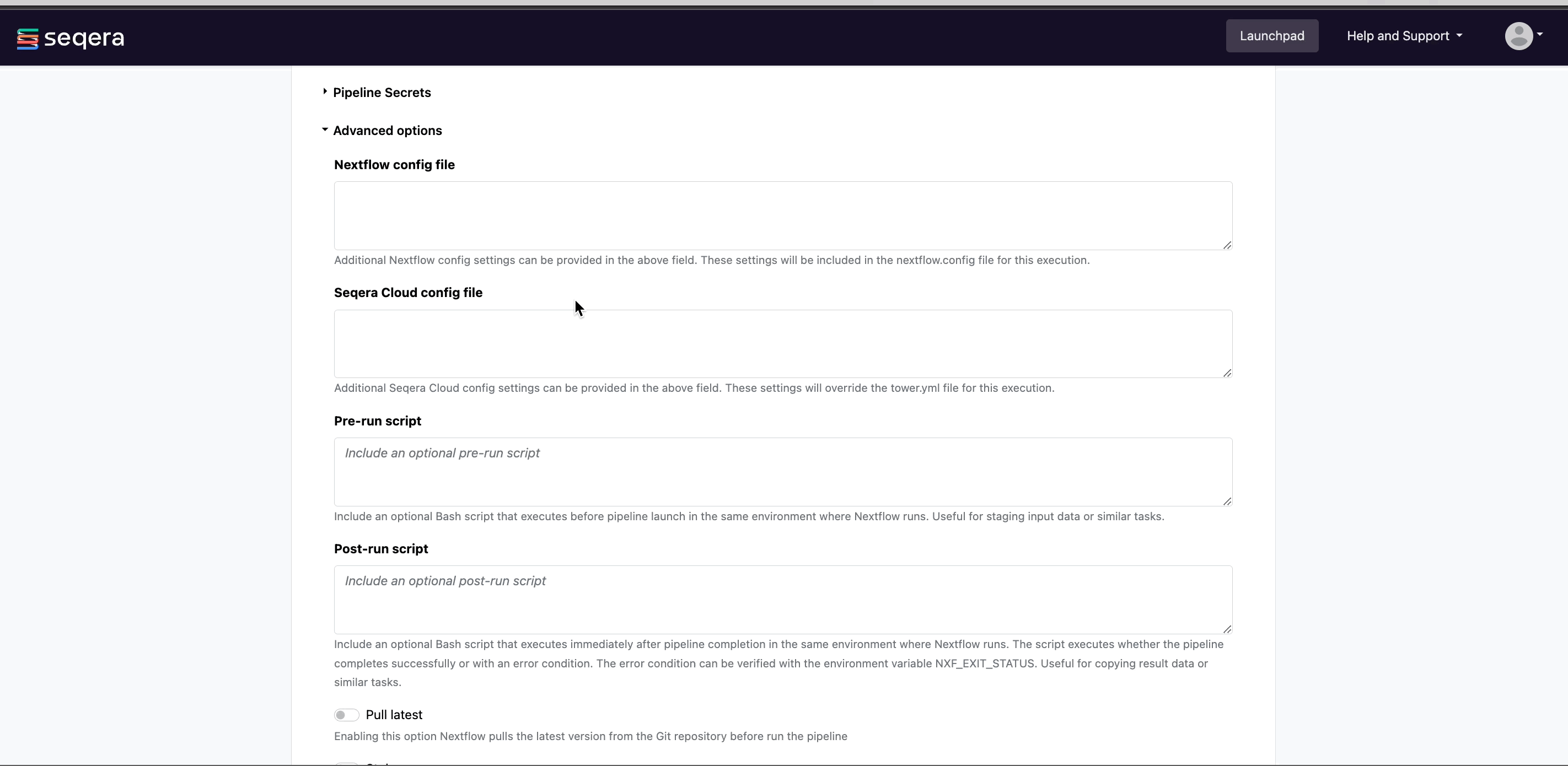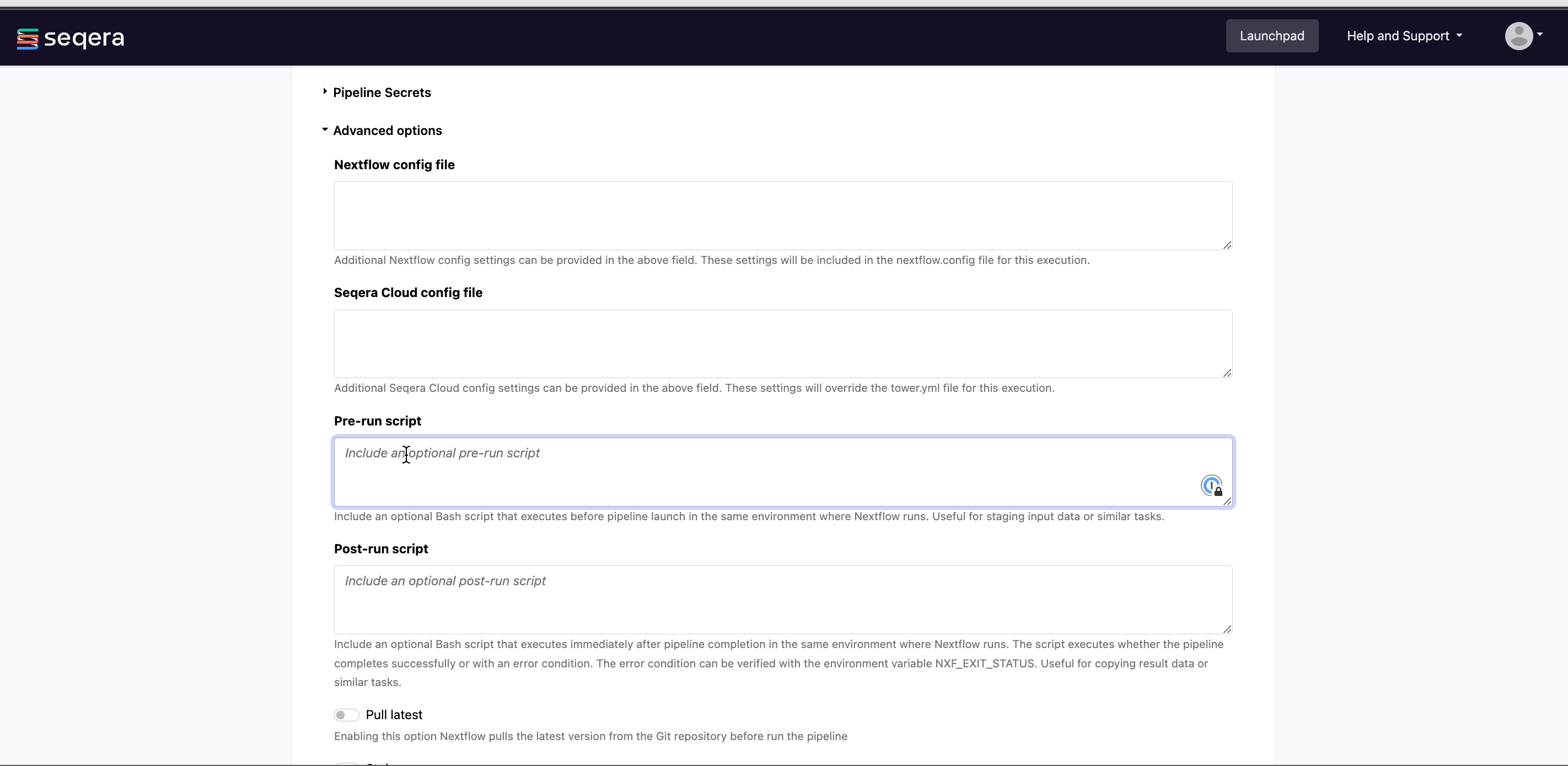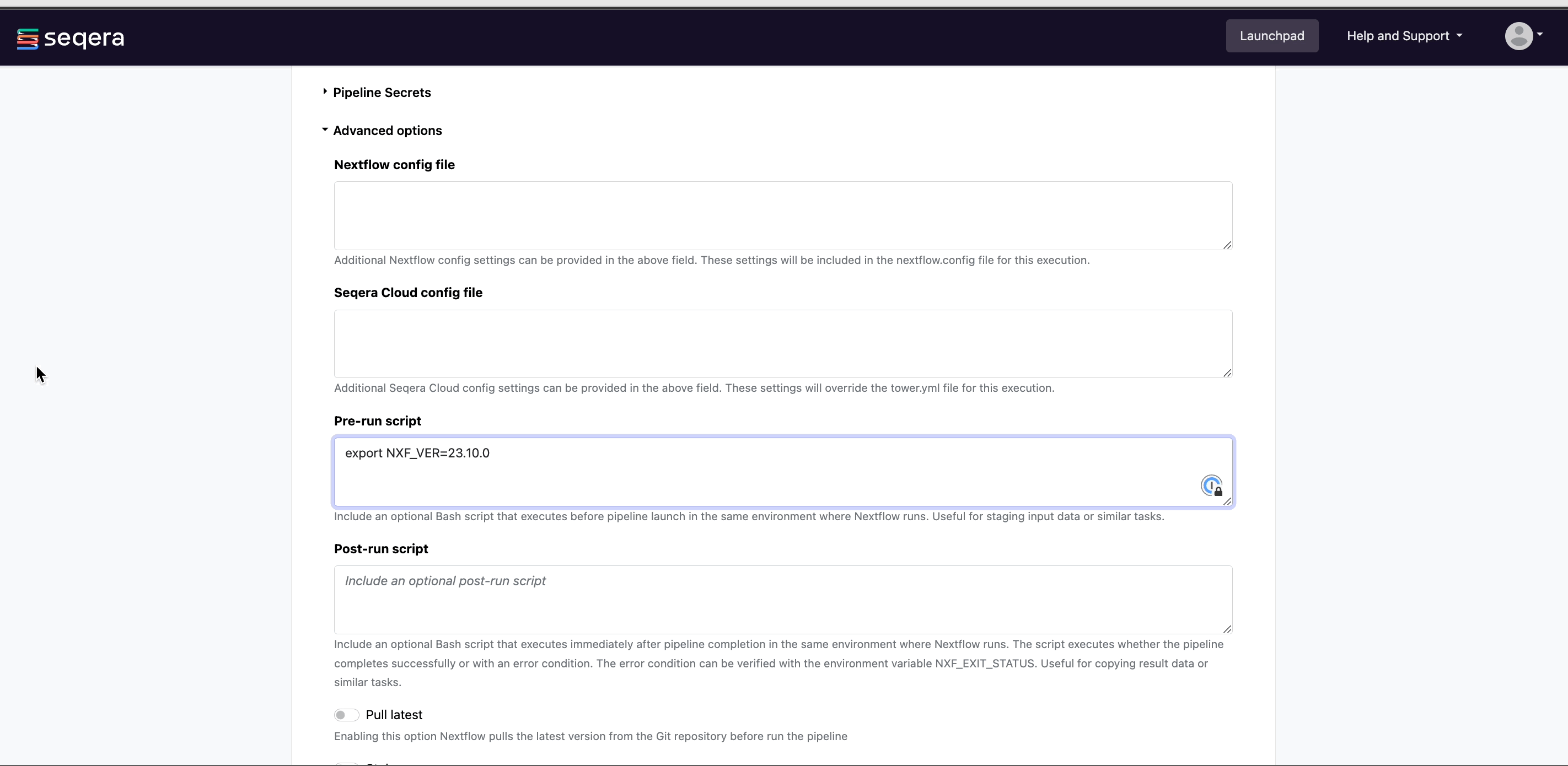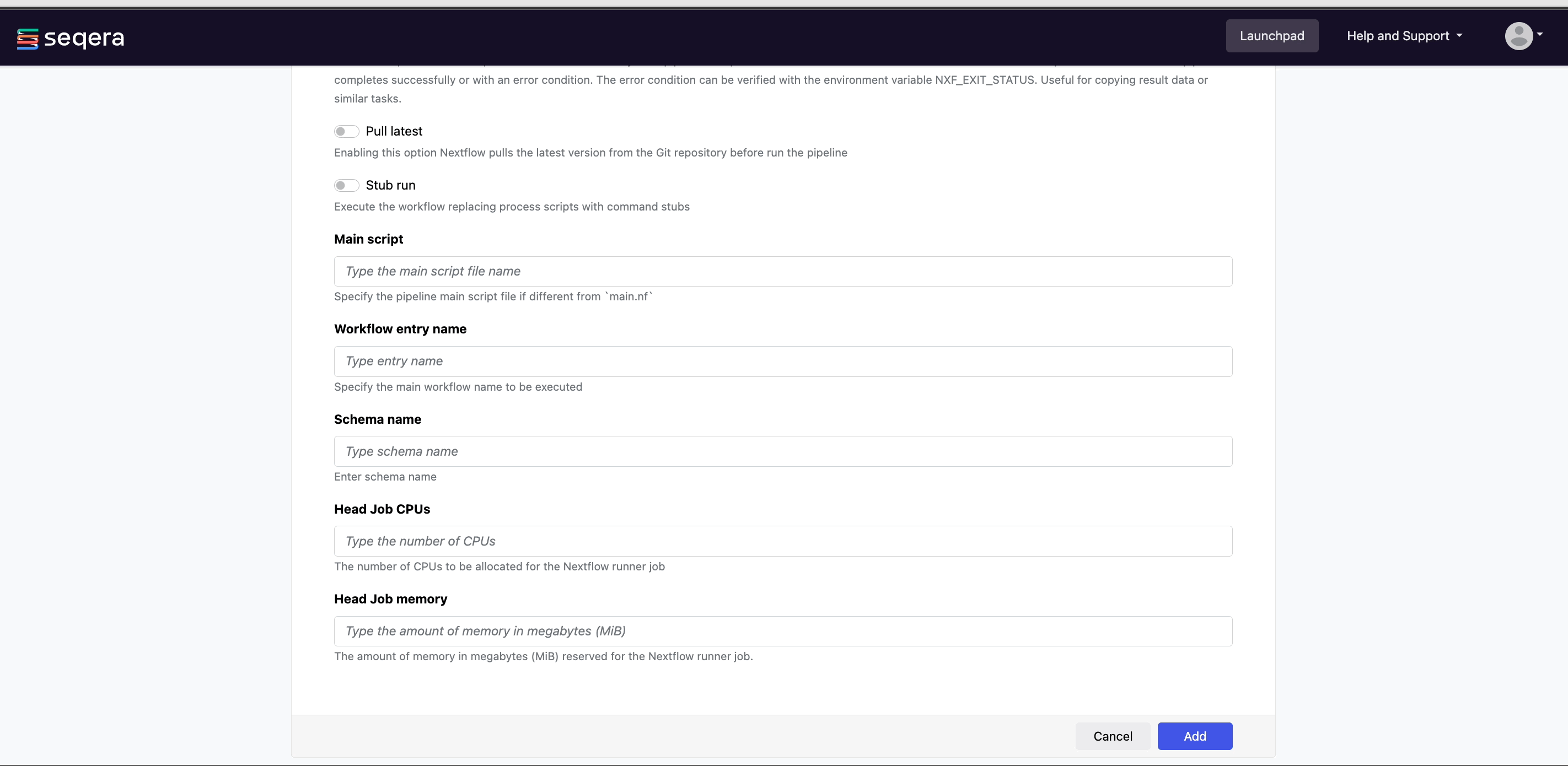
Pre-filled pipeline settings (such as compute environment, config profiles, and pipeline parameters) can be overridden during pipeline launch by workspace participants with the necessary permissions.
After you have populated the appropriate fields, select Add. nf-core/rnaseq is now available for workspace participants to launch in the preconfigured compute environment.